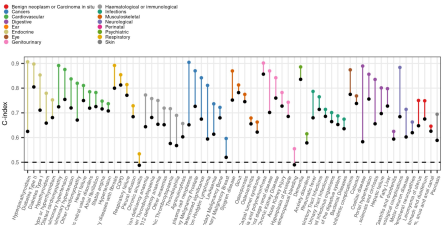
Proteomic signatures improve risk prediction for common and rare diseases
We developed sparse protein signatures to predict 10-year incidence of 218 common and rare diseases. Protein signatures significantly improved predictive performance over and above standard clinical risk factor for 67 diseases. For 52 diseases protein signatures further outperformed clinical biomarkers. Explore our results here.
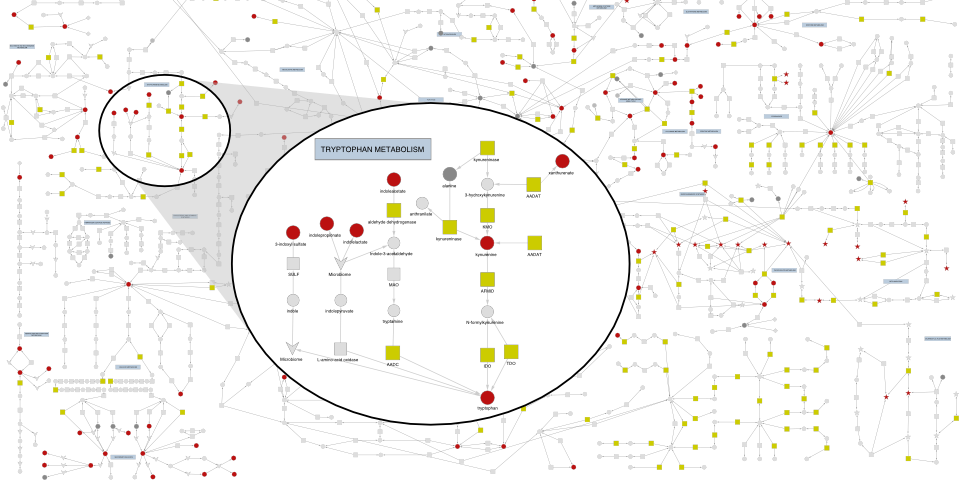
Rare and common genetic determinants of metabolic individuality
We studied the genetic architecture of the human plasma metabolome using 913 metabolites assayed in 19,994 individuals. This study identified 2,599 variant–metabolite associations within 330 genomic regions, with rare variants (minor allele frequency ≤ 1%) explaining 9.4% of associations. Summary statistics and pathway visualizations can be explored here.
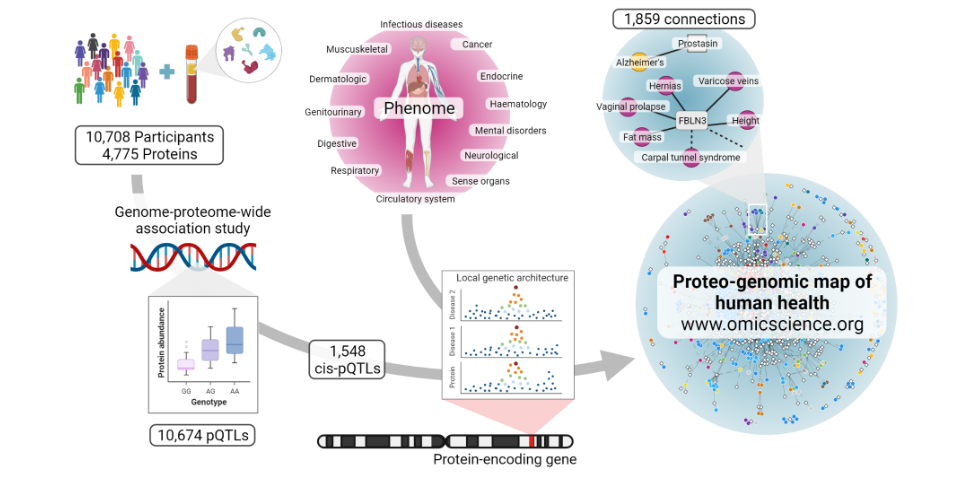
Mapping the proteo-genomic convergence of human diseases
We performed genome-proteome-wide association analysis for 4,775 human plasma proteins assayed by the SomaScan v4 platform among over 10,000 individuals, identifying 2,584 genomic regions associated with at least one out of 3,892 protein targets. Explore our results here.
Image generated using BioRender.
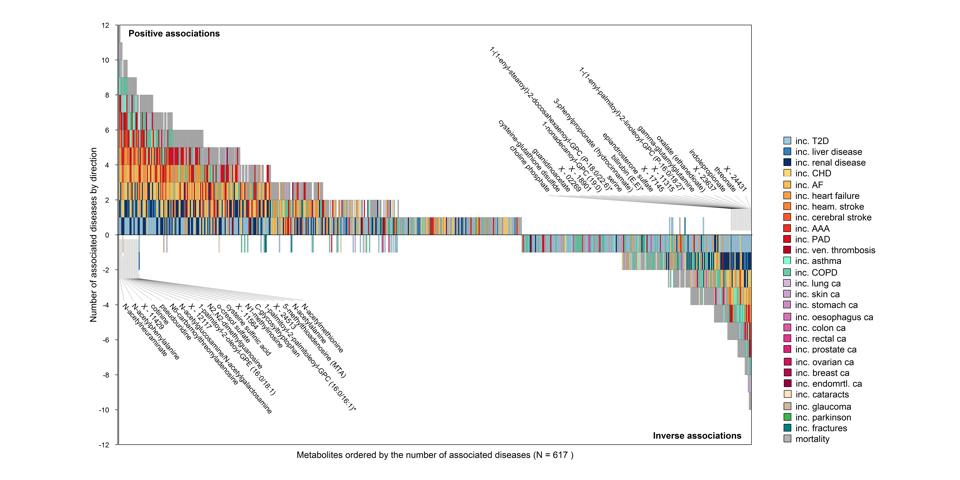
Shared and distinct metabolite profiles of incident diseases
Investigating the association of 1,014 blood metabolites with 27 incident diseases, all-cause mortality and disease multimorbidity in more than 11,000 participants, we identified shared and distinct pathways to non-communicable diseases and multimorbidity. We provide a comprehensive catalogue of risk factor - metabolite - disease associations by integrating data from over 50 modifiable clinical and other risk factors. Explore our results here.
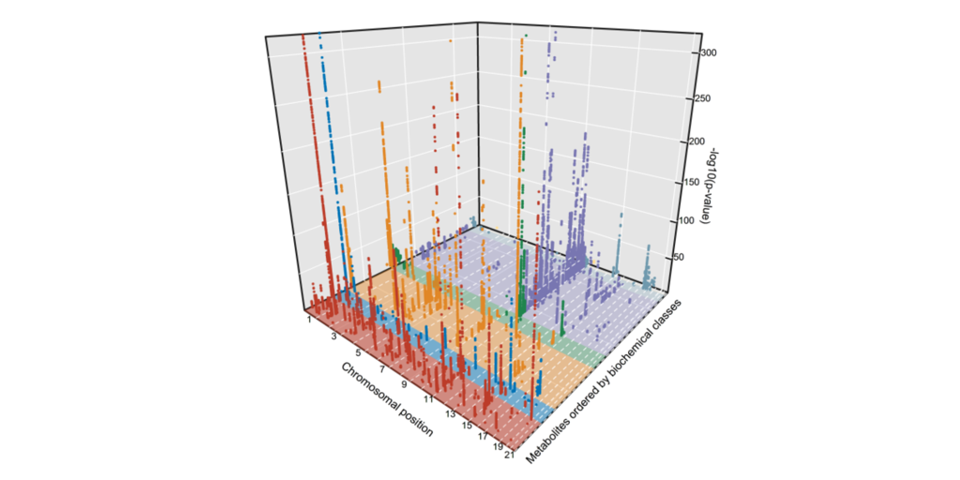
Cross-platform genetic discovery of metabolism products
In our cross-platform, genome-wide meta-analyses of 174 blood metabolites across six cohorts and including over 80,000 participants we identified 362 genome-wide significant associations and provide examples of the influence of metabolite-associated variants on human health. Summary statistics (meta-analyses and individual studies) are available here.
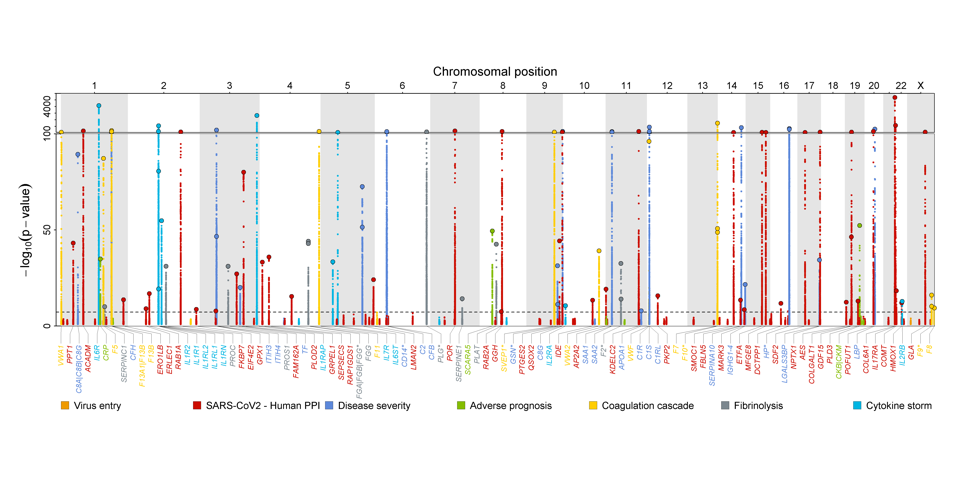
Genetic architecture of host proteins interacting with SARS-CoV-2
We integrated genomic and proteomic data from over 10,000 individuals to characterize the genetic architecture of host proteins reported to interact with SARS-CoV-2 or to be related to virus entry, the host hyper-immune or procoagulant response, or severity of COVID-19. Explore our results here.
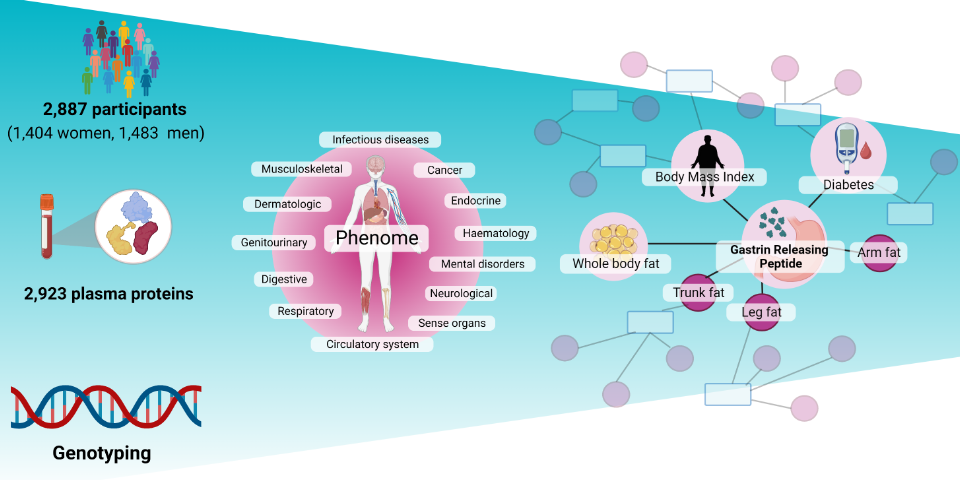
Proteogenomic links to human metabolic diseases
We conducted a cis-focused proteogenomic analysis of 2,923 plasma proteins measured in 1,180 individuals using the antibody-based Olink Explore 1536 and Explore Expansion assays. We identified a total of 1,553 cis-pQTLs (256 unreported) for 914 unique protein targets. Download our results here.